Photosynthesis, plants and environment
We are interested in the biogenesis of chloroplasts and their photosynthetic machinery. We aim to elucidate the regulatory networks underlying the build-up and wish to understand how plants integrate environmental fluctuations in order to maintain stable photosynthetic efficiency also under adverse conditions, an important aspect highly relevant to expected issues connected to the climate change.
Chloroplasts, light, life
Photosynthesis uses a complex apparatus of proteins, pigments and lipids to capture the light energy from sun and converts it into chemically usable energy. In this process, carbon dioxide from the air is assimilated and incorporated into sugar molecules. At the same time, the process releases oxygen and thus determines the gas composition of the atmosphere.
Only plants, algae and some bacteria are capable of photosynthesis. Plants and algae have special intracellular organelles for this purpose, the chloroplasts, in which photosynthesis takes place. Evolutionarily, chloroplasts emerged from the endosymbiotic uptake of a photosynthetic bacterium into a unicellular heterotrophic eukaryote around 1.2 Mya. Therefore, chloroplasts still exhibit several prokaryotic features including an own genome (called plastome) and a complete gene expression machinery. In flowering plants chloroplasts are inherited as an undifferentiated, non-photosynthetic proplastid. Germinating seedlings thus depend on their storage energies to grow and aim to reach the light as quick as possible to become photosynthetic. Once the first leaves are illuminated the light triggers a developmental cascade called photomorphogenesis eventually leading to a photosynthetically active green seedling.
The formation of the photosynthetic apparatus in chloroplasts in of plants cells in turn requires the expression of photosynthesis genes encoded in the plastid genome and the cell nucleus. The build-up of photosynthesis thus includes light as a trigger as well as a complex molecular regulation that controls the provision of all components of the photosynthetic apparatus. One key element of this regulatory network is the chloroplast RNA polymerase that is responsible for the proper expression of chloroplast photosynthesis genes. It integrates developmental and environmental signals eventually leading to a photosynthetically active seedling.
Photosynthesis
Photosynthetic machinery
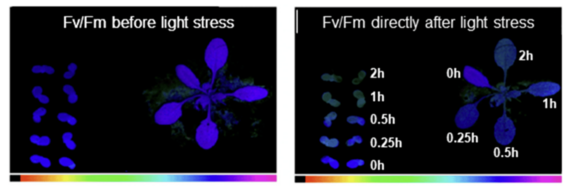
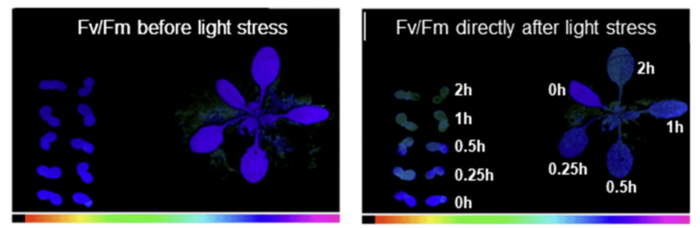
The structural and functional basics of plant photosynthesis have been extensively studied at cellular and molecular level. At present the construction of the photosynthetic complexes within the thylakoid membrane system of plant chloroplasts is well understood and the position and coordination of proteins, pigments, lipids and co-factors is described even at atomic level. However, the photosynthetic machinery is not static, but rather represents a highly dynamic arrangement of complexes that change position and association depending on environmental conditions. The photosynthetic machinery, therefore, performs two functions:
- The primary function of photosynthesis is the harnessing of sun light and the subsequent conversion into chemical energy.
- The second, less obvious function is the sensing of the environment. The photosynthetic process is highly susceptible to changes in light, temperature or water availibility and further is highly affected by pathogen or herbivore attack.
We are especially interested in the sensor function of photosynthesis as it regulates many plant-internal processes such as metabolism and gene expression. The sensor function, however, controls also photosynthetic function by regulatory feedback loops and acclimates photosynthesis to environmental conditions by structural rearrangements of the photosynthetic apparatus.
One key regulatory point in photosynthesis is the redox state of the intermittent plastoquinone (PQ) pool that electrochemically connects photosystem II (PSII) and the cytochrome b6f complex. In the lab we manipulate the PQ redox state by preferential excitation of PSII or PSI using artificial light sources that create excitation imbalances between the two photosystems. This way of experimental variation in illumination mimics light quality gradients typically found in dense plant stands (such as crop fields or natural canopies) or under industrial farming conditions such as green houses or vertical farming arrangements.
Molecular properties of duckweed photosynthesis
We aim to understand how photosynthesis acclimates to light quality gradients in natural and artificial surroundings and how this can be exploited to improve agri- and horticultural productivity. Most of our research has been performed with model organisms such as Arabidopsis, Sinapis or Nicotiana. We, however, are also interested in the study of novel plant systems that are of interest for biotechnology and biomass production. To this end we study photosynthesis in duckweeds (Lemnaceae), a family of aquatic plants capable of exponential growth if grown under appropriate conditions. Duckweeds are freshwater plants that can be found ubiquitiously on planet Earth apart from polar habitates. They multiply by vegetative fission and can exhibit tremendous growth rates. In recent years they are heavily investigated for their potential bioeconomic impact in food, feed, biomass and biotechnological production and application. We study the molecular properties of duckweed photosynthesis with respect to structure and physiological acclimation.
Chloroplast RNA Polymerase
PAPs: plastid encoded RNA polymerase associated proteins



Chloroplast transcription is performed by two RNA polymerases, a phage-type nuclear-encoded RNA polymerase (NEP) and a cyanobacteria-type plastid encoded RNA polymerase (PEP). The core subunits of PEP are encoded by rpo the genes located on the plastome. These genes (and a few others) are transcribed by NEP. PEP transcribes most of the other genes especially those for photosynthesis and is absolutely essential for biogenesis of chloroplasts and the photosynthetic apparatus. Intriguingly, the PEP complex is expanded during photomorphogenesis by the association of at least 12 additional protein subunits. These PEP-associated proteins (PAPs) provide a number of further enzymatic properties to PEP, the purpose of which is however, largely not understood.
PAPs represent RNAP additions for which no orthologues can be found in any other prokaryotic or eukaryotic multisubunit RNAP. Thus, they are chloroplast-specific. We are keen to understand their specific roles and their evolution. Functions of PAPs are largely enigmatic and only protein domain predictions and a few in vitro data with recombinant proteins do exist. According to this PAPs exert major functions in DNA/RNA metabolism, redox regulation various enzymatic activities and yet unknown functions. Regardless of their diversity they share some common features. Most prominent is the phenotype of PAP inactivation mutants that are all pigment-deficient displaying albino – pale-green phenotypes. Further, several PAPs appear to be dually localized to plastids and nucleus. Current data suggest that they play important coordinative roles in retrograde signalling and photomorphogenesis, two hot topics in the field of molecular regulation of plant development.
Retrograde signalling
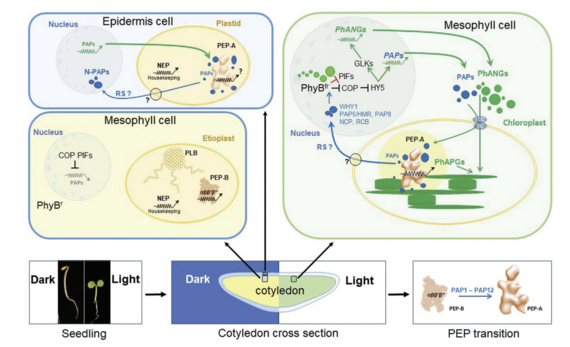
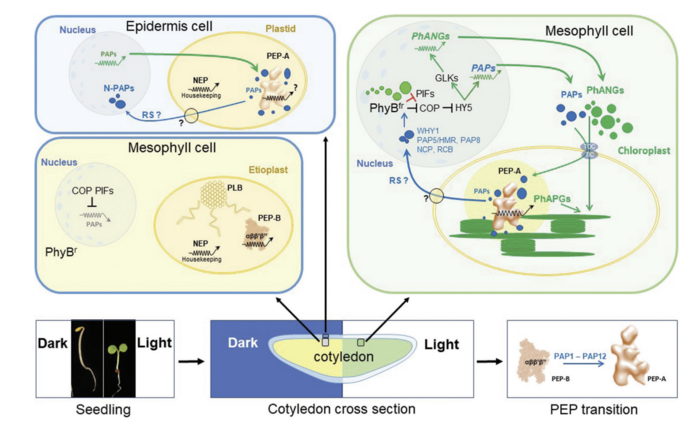
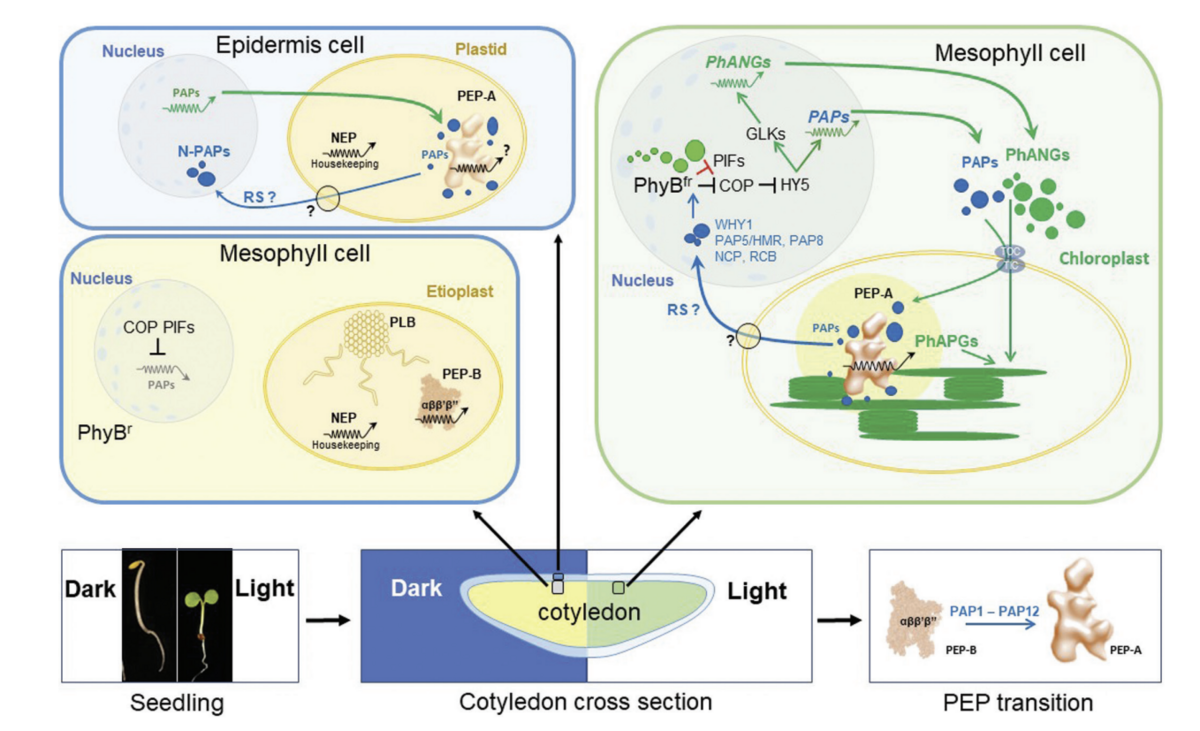
Chloroplasts act not only as central biosynthetic platforms in plant cells, but also as sensors for a multiplicity of environmental and internal developmental cues. The photosynthetic process, in particular, is highly sensitive to abiotic and biotic stresses and provides a number of signals towards the nucleus reporting stresses. These signals initiate the expression of genes that trigger the action of compensatory responses with the aim to replenish deleterious stress effects. Since these signals originate mostly from disturbances in the balance of photosynthesis-associated redox processes they are summarised under the term „operational retrograde control“.
The initial steps of chloroplast biogenesis during seedling development are triggered by the nucleo-cytosolic phytochrome system that reports important developmental signals from the nucleus towards the emerging chloroplasts. This signalling route was termed „anterograde signalling“. Any disturbance in the normal development of chloroplasts, however, is reported back to the nucleus informing about deficiencies in in organelle build-up. These signals are summarised under the term „biogenic retrograde control“. For a critical discussion please read Liebers et al 2022 JXB.
We study both types of retrograde signalling in depth as they are intrisically connected to our two research pillars:
- Photosynthetic redox signals generated by environmental fluctuations do not only acclimate the photosynthetic apparatus but also influence nuclear and cytosolic processes.
- Several PAPs are dually localized proteins residing both in nucleus and plastids. PAP5 and PAP8 have been shown to interact with the phytochrome system and to provide a novel means of biogenic retrograde signalling.
Research grants
Current grants
Contact


30419 Hannover




